multiTimelineUFO <- read_csv("data/multiTimelineUFO.csv", skip = 1)
trends_ufo <- multiTimelineUFO %>%
janitor::clean_names() %>%
mutate(month = yearmonth(month)) %>%
rename(hits = ufo_united_states) %>%
as_tsibble(index = month)Activity49
Intro & Why Organization Matters
Today we’ll learn professional report structuring in R Markdown. Key goals:
Clean main text with
>hidden code>Appendix for reproducibility
>Optimized figures/tables with captions
Logical
>markdown headings”
Basic Chunk Management
```{r setup, include=FALSE}
# Libraries, data load, functions here
knitr::opts_chunk$set(echo=FALSE, message=FALSE)
```Add a figure caption
```{r fig_main, fig.cap="Main time series plot"}
p_main # Plot object from your code
```Key points:
>include=FALSE hides setup code completely
>echo=FALSE shows output without code
- Always use
>fig.cap for figures
Advanced Figure/Table Formatting
```{r model_table, results='asis'}
knitr::kable(model_comparison, caption="Model accuracy comparison")
``````{r decomposition, fig.cap="STL decomposition", fig.width=8, fig.height=5}
p_stl # Your decomposition plot
```Table/figures:
>kable(caption = “A nice caption”) for tables with captions
>fig.width/height adjustments
>results=‘asis’ for proper table rendering
Residual Diagnostics Section
Diagnostics
```{r residuals, fig.cap="ARIMA residuals", fig.subcap=c("TS", "ACF", "Hist")}
arima_fit %>% gg_tsresiduals()
```Emphasize:
>Multi-panel figures with subcaptions
- Consistent
>theme_minimal usage
- Interpretation in text near figures
Appendix Creation
```{r appendix_code, echo=TRUE}
# Smaller chunks, with informative comments
trends_ufo %>% autoplot() # Original plotting code
```Critical skills
>eval=FALSE prevents duplicate computation
- Organized
>code documentation
Final Polish
- Show
>heading hierarchy:# Section>## Subsection
- Demonstrate
>global chunk options in setup
Cheatsheet:
Essential Chunk Options:
• echo=FALSE - Hide code
• include=FALSE - Run but hide everything
• fig.cap="Text" - Figure captions
• results='asis' - For tables
• fig.width=8 - Control plot size
• eval=FALSE - Show code without runningClass Activity
Create a well-organized skeletal report analyzing UFO search Trends in US using R Markdown. Your report must:
- Separate code execution from presentation using include=FALSE for setup/data chunks and echo=FALSE for result displays
- Show all figures/tables in the main text with descriptive captions (fig.cap/kable(caption=))
- Include full reproducible code in an Appendix using eval=FALSE, echo=TRUE
- Compare models professionally with knitr::kable() formatted tables
- Structure content clearly under these headings: Introduction, Results (Trends/Models), Diagnostics, Appendix
Practice report-writing skills by:
- Keeping the main text code-free while showing visualizations
- Using chunk labels consistently (e.g.,
fig_main,tbl_models)
- Cross-referencing appendix code to main text elements
- Optimizing figure sizes via
fig.width/height
Introduction
Methodology
Results
Main Trends
trends_ufo %>% autoplot(hits) +
labs(y = "Relative Interest (0-100)", x = "Month") +
theme_minimal()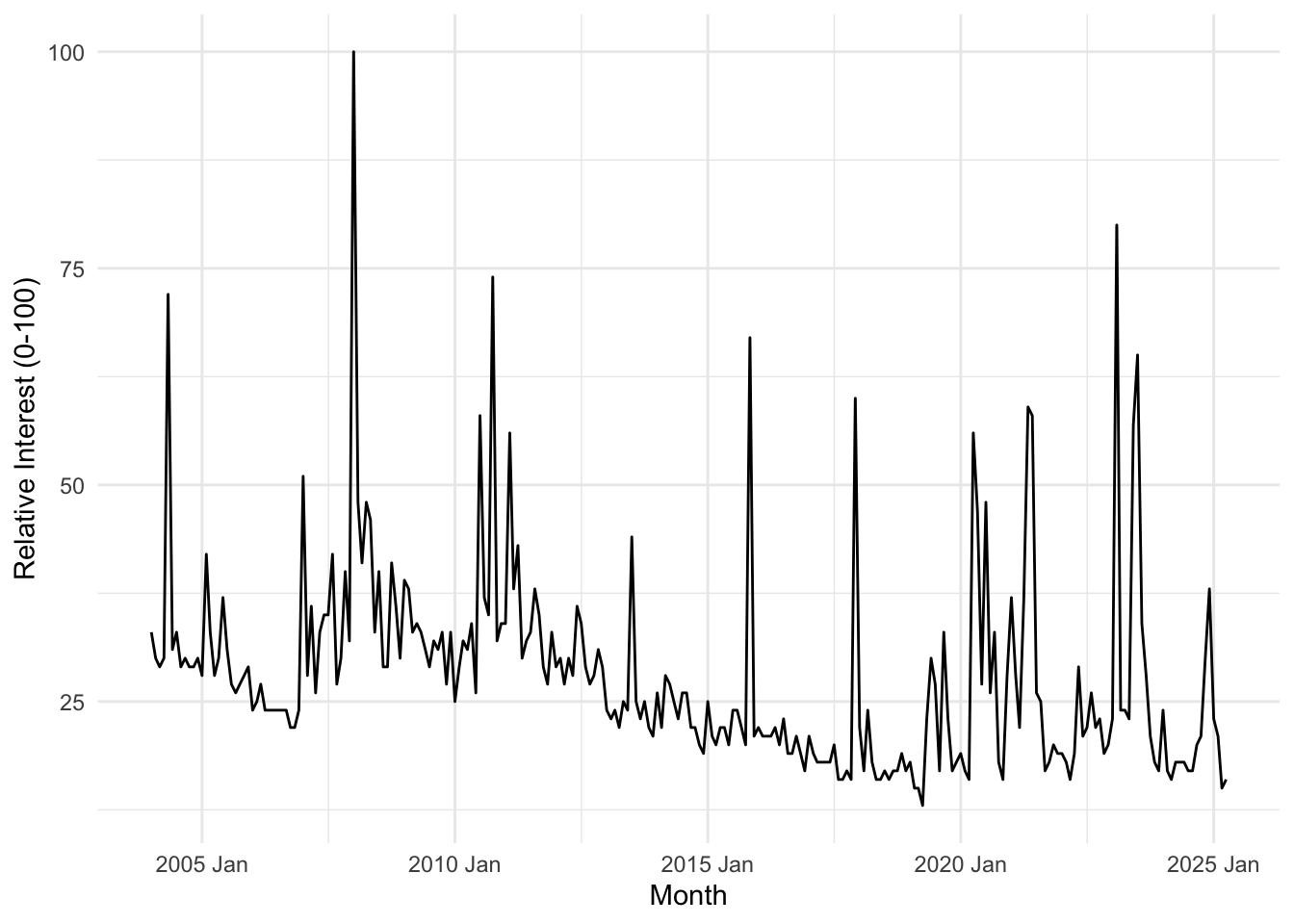
Model Comparisons
arima_fit <- trends_ufo %>% model(ARIMA(hits))
lambda_est <- trends_ufo %>% features(hits, guerrero) %>% pull(lambda_guerrero)
arima_fit_bc <- trends_ufo %>% model(ARIMA(box_cox(hits, lambda_est)))
ets_fit_bc <- trends_ufo %>% model(ETS(box_cox(hits, lambda_est)))model_comparison <- accuracy(arima_fit_bc) %>%
bind_rows(accuracy(ets_fit_bc)) %>%
bind_rows(accuracy(arima_fit))
knitr::kable(model_comparison, caption = "Model Accuracy Metrics Comparison")| .model | .type | ME | RMSE | MAE | MPE | MAPE | MASE | RMSSE | ACF1 |
|---|---|---|---|---|---|---|---|---|---|
| ARIMA(box_cox(hits, lambda_est)) | Training | 0.6205578 | 10.58485 | 5.845902 | -5.234272 | 18.13860 | 0.6410787 | 0.6970692 | -0.0440546 |
| ETS(box_cox(hits, lambda_est)) | Training | 0.6623999 | 10.65716 | 5.843008 | -4.918688 | 18.20439 | 0.6407613 | 0.7018310 | 0.0625724 |
| ARIMA(hits) | Training | -0.4361228 | 10.64533 | 6.422410 | -9.764741 | 21.20102 | 0.7043002 | 0.7010519 | -0.0077131 |
Diagnostics
ARIMA and ETS Residuals
arima_fit %>% gg_tsresiduals()
ets_fit_bc %>% gg_tsresiduals()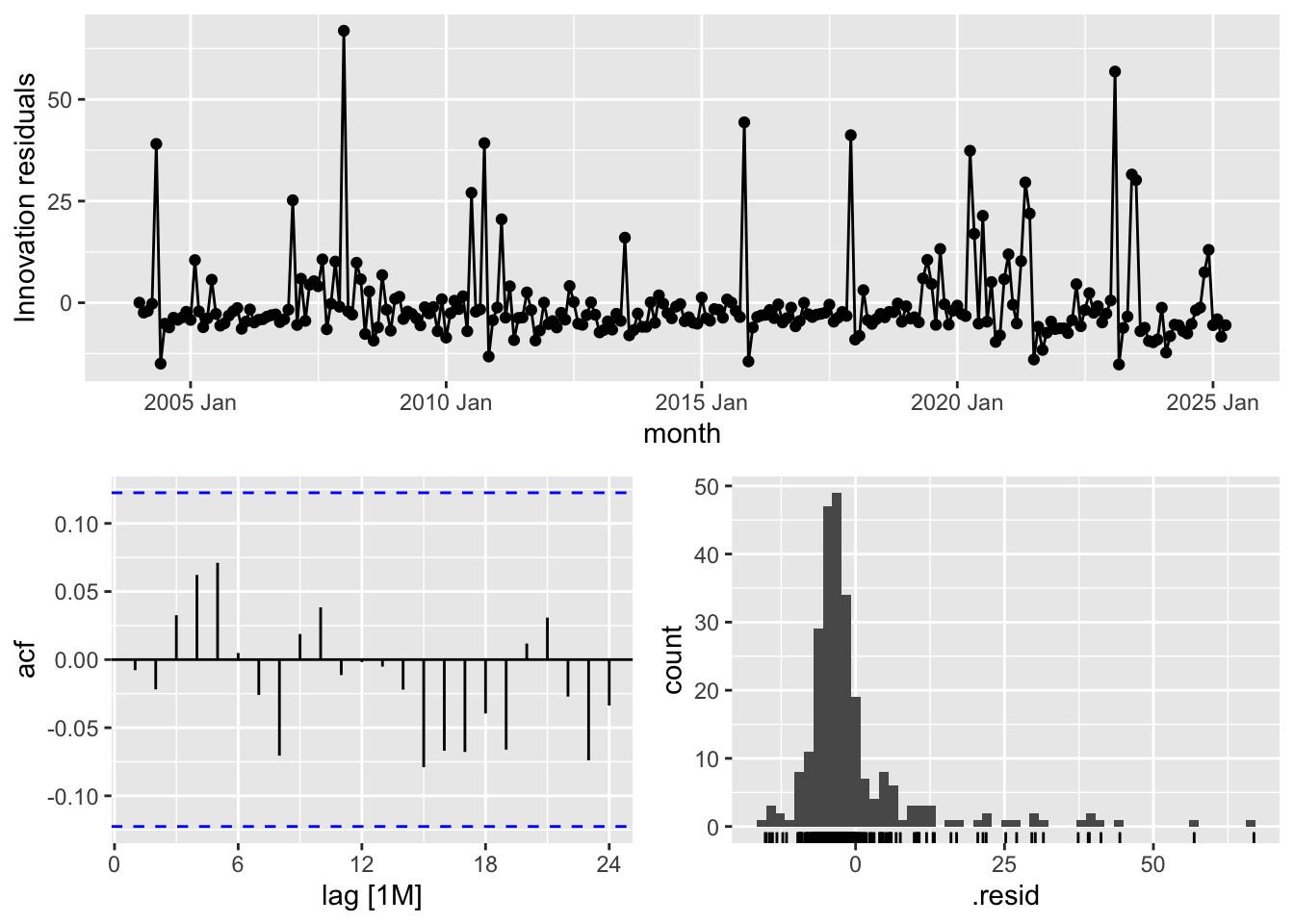
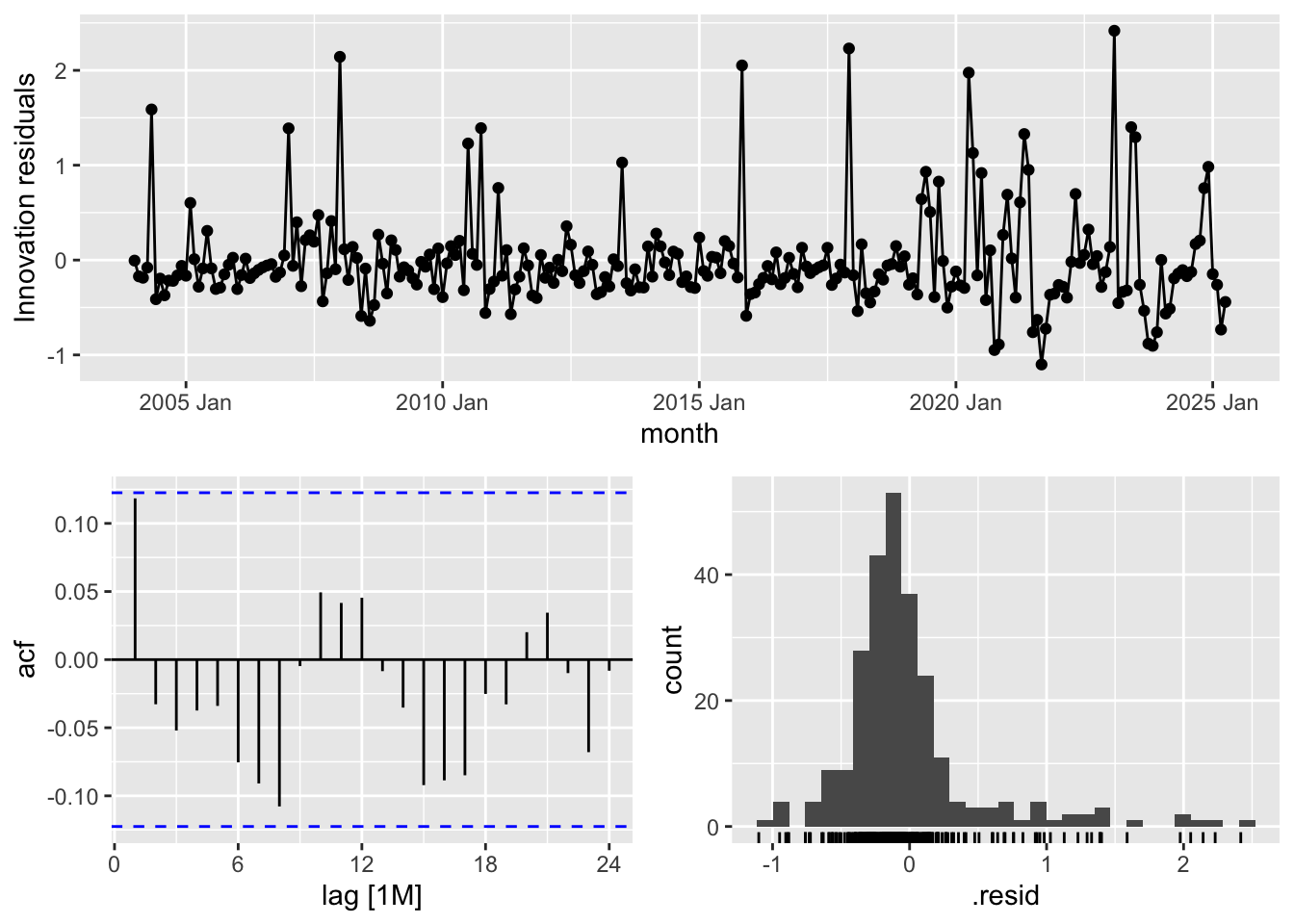
Residual Diagnostics for ARIMA and ETS Models