# Retrieve COVID-19 data for the United States and prepare a tsibble
covid_data <- covid19(verbose = FALSE) %>%
filter(administrative_area_level_1 == "United States") %>%
mutate(date = ymd(date)) %>%
as_tsibble(index = date) %>%
select(date, confirmed, vaccines) %>%
mutate(vaccines = as.integer(vaccines)) %>%
drop_na()Activity16
COVID Data Analysis and Granger causality test
Objective
Investigate whether changes in COVID-19 vaccination numbers help predict changes in confirmed cases using a Granger causality test.
# Compute daily changes in confirmed cases and vaccinations
covid_data <- covid_data %>%
mutate(dConfirmed = difference(confirmed),
dVacc = difference(vaccines)) %>%
drop_na()# Plot daily changes in confirmed cases and vaccinations
covid_data %>%
autoplot(vars(dConfirmed, dVacc)) +
labs(title = "Daily Changes: Confirmed Cases & Vaccinations", x = "Date")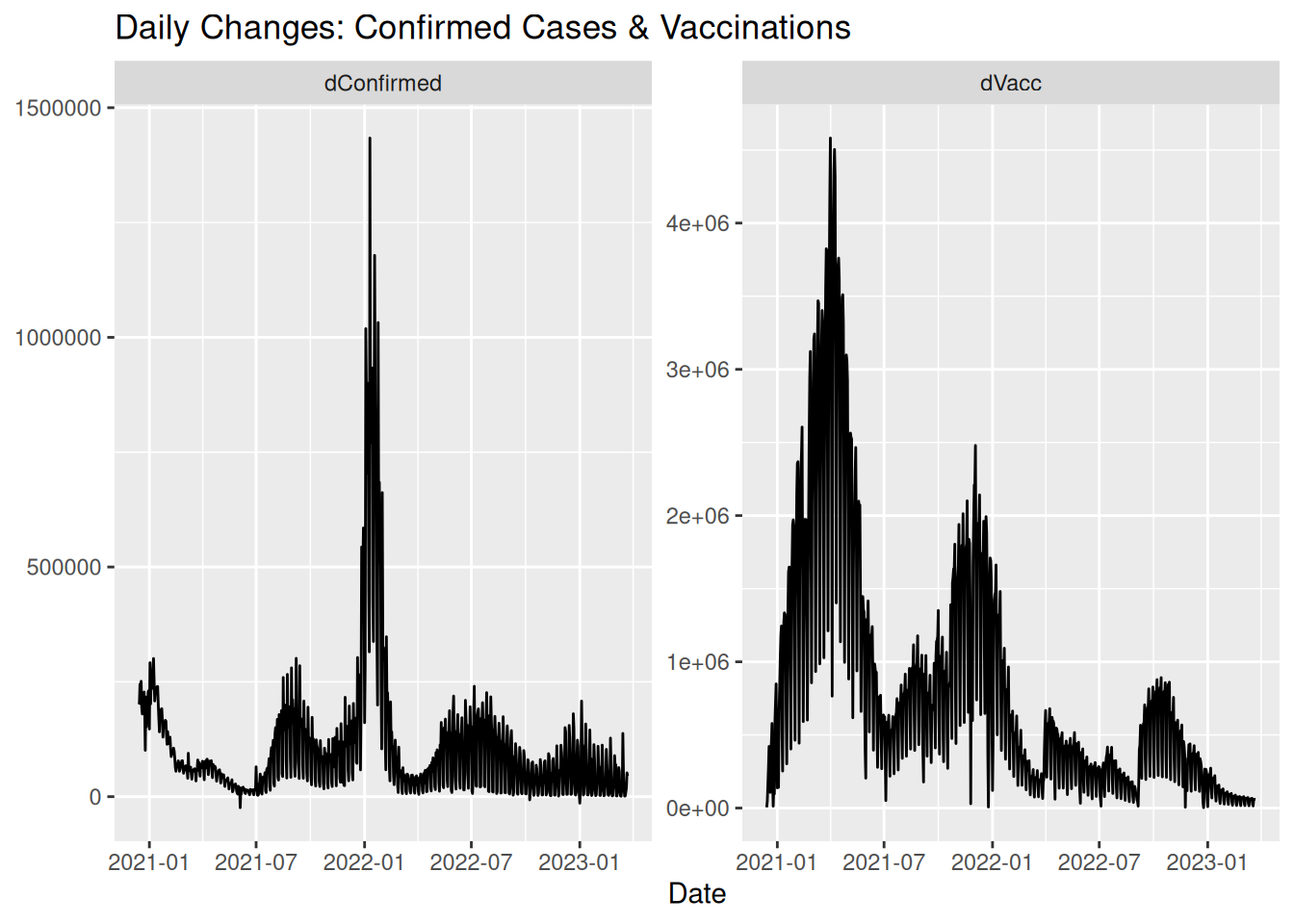
Granger Causality Test: Theory & Implementation
The Granger causality test checks whether past values of one variable (here, dVacc) provide statistically significant information for predicting another variable (dConfirmed).
Implications and Uses:
- A significant result (p-value < 0.05) suggests that changes in vaccinations Granger-cause changes in confirmed cases.
- This does not imply true causality, but indicates that past vaccination data improves forecast accuracy for confirmed cases.
- Such insights can help inform public health policies by highlighting the predictive value of vaccination trends.
P-value Interpretation:
- p < 0.05: Reject the null hypothesis. Past vaccination changes significantly improve prediction of confirmed cases.
- p ≥ 0.05: Fail to reject the null hypothesis. Vaccination changes do not add significant predictive power.
VAR Modeling and IRF Computation
# Test if changes in vaccinations (dVacc) Granger-cause changes in confirmed cases (dConfirmed) using a lag of 1.
model_full <- lm(dConfirmed ~ lag(dVacc, 1), data = covid_data)
model_restricted <- lm(dConfirmed ~ 1, data = covid_data)
F_stat <- ((deviance(model_restricted) - deviance(model_full)) / 1) /
(deviance(model_full) / (nrow(covid_data) - 2 - 1))
df1 <- 1
df2 <- nrow(covid_data) - 2 - 1
p_value <- pf(F_stat, df1, df2, lower.tail = FALSE)
p_value[1] 0.260462Lab Activity: Exploring Granger Causality with Different Lags
Task: Using the covid_data, perform Granger causality tests with lag orders 1, 2, 3, 5, and 10.
For each lag, compute the p-value to assess whether lagged vaccination changes improve the prediction of confirmed cases. Answer the following:
- How do the p-values change with different lag orders?
- Which lag order appears most appropriate for capturing the predictive relationship?
Write a brief discussion on how these findings might inform public health policy.