# Retrieve COVID-19 data for the United States and prepare a tsibble
# Get cumulative COVID metrics
covid_raw <- covid19(country = "US", level = 1, verbose = FALSE) %>%
as_tsibble(index = date) %>%
select(date, confirmed, people_vaccinated) %>%
rename(vaccines = people_vaccinated)Activity41
Structural Breaks in COVID-19 Data
The COVID-19 pandemic created distinct epidemiological regimes where parameters governing case growth changed fundamentally. Identifying these structural breaks through vaccine rollouts, policy changes, and variant emergence helps quantify intervention effectiveness and improve forecasting. This case study demonstrates how to detect and model regime shifts using FDA emergency vaccine authorization (Jan 2021) as a natural experiment.
\[ \begin{aligned} \text{Confirmed}_t &= \underbrace{\phi_0 + \phi_1 \text{Confirmed}_{t-1}}_{\text{Autoregressive component}} + \underbrace{\beta \text{Vaccines}_t}_{\text{Current vaccines}} + \underbrace{\gamma D_t}_{\text{Break effect}} + \epsilon_t \\ D_t &= \begin{cases} 1 & \text{if } t \geq \tau \ (\text{post-break}) \\ 0 & \text{otherwise} \end{cases} \\ \epsilon_t &\sim \text{WN}(0, \sigma^2) \end{aligned} \]
Data Preparation
We first convert raw cumulative COVID-19 data to new weekly cases/vaccinations using lagged differences. Log-transformation stabilizes variance while preserving relative changes. Weekly aggregation reduces daily reporting noise (e.g., weekend under-reporting).
# Convert to NEW weekly cases
covid_weekly <- covid_raw %>%
mutate(across(c(confirmed, vaccines), ~. - lag(.))) %>% # Calculate daily changes
index_by(week = yearweek(date)) %>%
summarize(
cases = sum(confirmed, na.rm = TRUE),
vaccines = sum(vaccines, na.rm = TRUE)
) %>%
filter(cases > 0, vaccines >= 0) %>%
mutate(across(c(cases, vaccines), ~ log(. + 1))) Activity 1: Visual Break Identification
covid_weekly %>% as_tibble() %>%
pivot_longer(-week) %>%
ggplot(aes(week, value)) +
geom_line() +
geom_vline(
xintercept = as.Date(yearweek("2021-01-11")),
color = "red",
linetype = "dashed"
) +
facet_grid(name ~ ., scales = "free") +
labs(
title = "U.S. COVID Trends with Vaccine Breakpoint Hypothesis",
subtitle = "Red line marks FDA emergency vaccine approval date"
)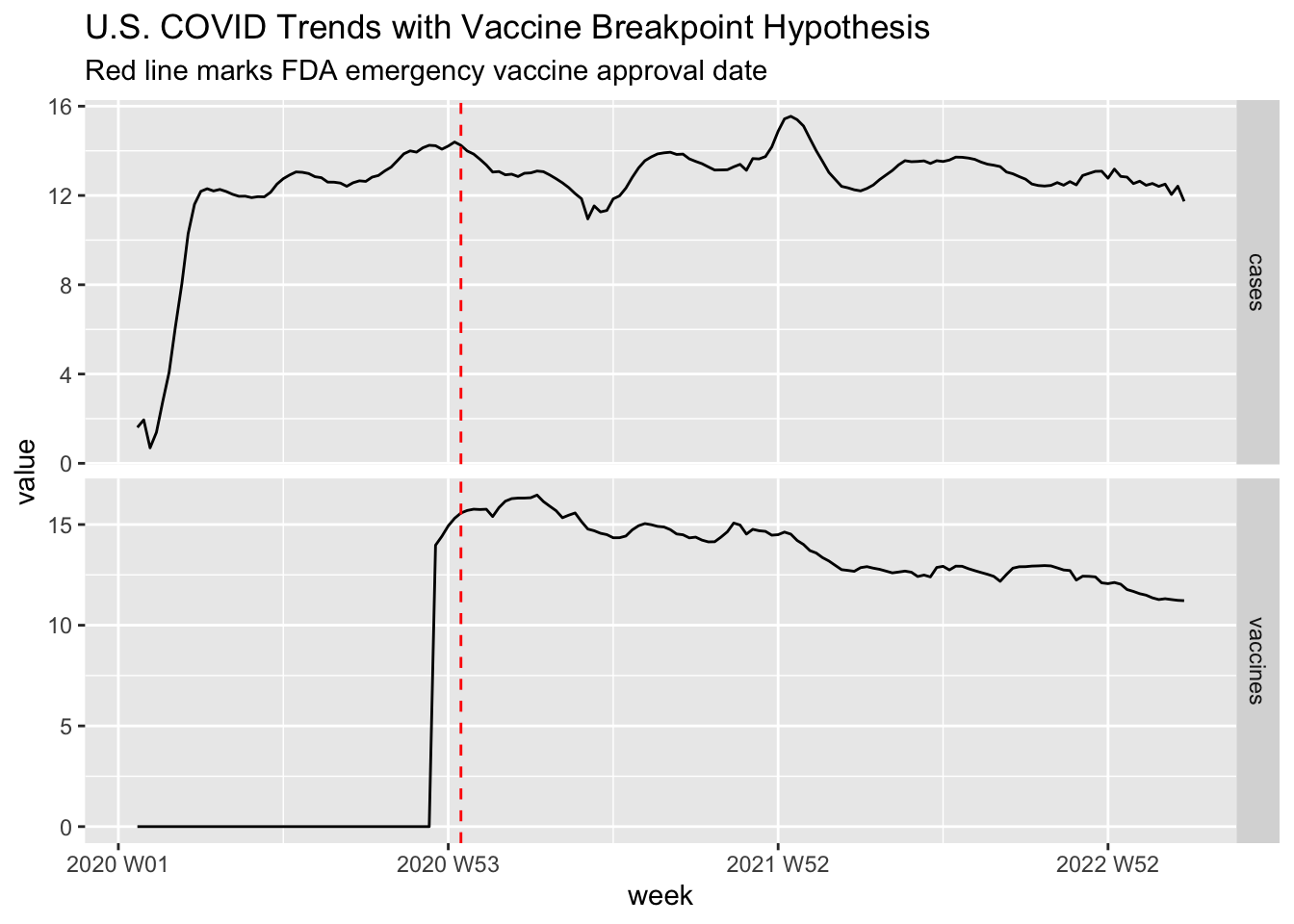
The vertical line marks January 2021 when vaccines became widely available. Visual analysis suggests a potential structural break - case growth deceleration post-vaccination despite similar transmission conditions.
Activity 2: Formal Breakpoint Test
library(strucchange)
bp_test <- Fstats(cases ~ vaccines, data = covid_weekly)
bp_cases <- breakpoints(cases ~ 1, data = covid_weekly, h = 0.15)
break_week <- covid_weekly$week[bp_cases$breakpoints]
# Create regime dummy
covid_weekly <- covid_weekly %>%
mutate(post_vax = (week >= break_week) %>% as.integer())Key Equation: Structural Break Model
\[ \begin{aligned} \log \left(\text { Cases }_t\right)&=\phi_0+\phi_1 \log \left(\text { Cases }_{t-1}\right)+\beta \log \left(\text { Vaccines }_t\right)+\gamma D_t+\epsilon_t \\ \text{where } D_t &= \begin{cases} 0 & \text{pre-vaccine} \\ 1 & \text{post-vaccine} \end{cases} \end{aligned} \] ## Activity 3: Modeling and Forecasting with Breaks
library(forecast)
# Convert to ts objects
cases_ts <- ts(covid_weekly$cases,
frequency = 52,
start = yearweek(min(covid_weekly$week)))
xreg_matrix <- covid_weekly %>% as_tibble() %>%
select(vaccines, post_vax) %>%
mutate(post_vax = as.numeric(post_vax)) %>%
data.matrix()
# Auto-ARIMAX
arimax_forecast <- auto.arima(
cases_ts,
xreg = xreg_matrix
)
arimax_forecast %>% summary()Series: cases_ts
Regression with ARIMA(4,0,2)(1,0,0)[52] errors
Coefficients:
ar1 ar2 ar3 ar4 ma1 ma2 sar1 intercept
2.1919 -1.0385 -0.6612 0.5042 -0.9009 0.2029 0.0401 11.2746
s.e. 0.1279 0.3349 0.3368 0.1235 0.1331 0.1815 0.1069 1.9063
vaccines post_vax
-0.0022 0.0500
s.e. 0.0172 0.2381
sigma^2 = 0.08649: log likelihood = -30.56
AIC=83.12 AICc=84.83 BIC=117.35
Training set error measures:
ME RMSE MAE MPE MAPE MASE
Training set 0.02989448 0.2850902 0.1778063 -0.6505711 3.342386 0.1153798
ACF1
Training set -0.01963548# Residual diagnostics
arimax_forecast %>%
checkresiduals()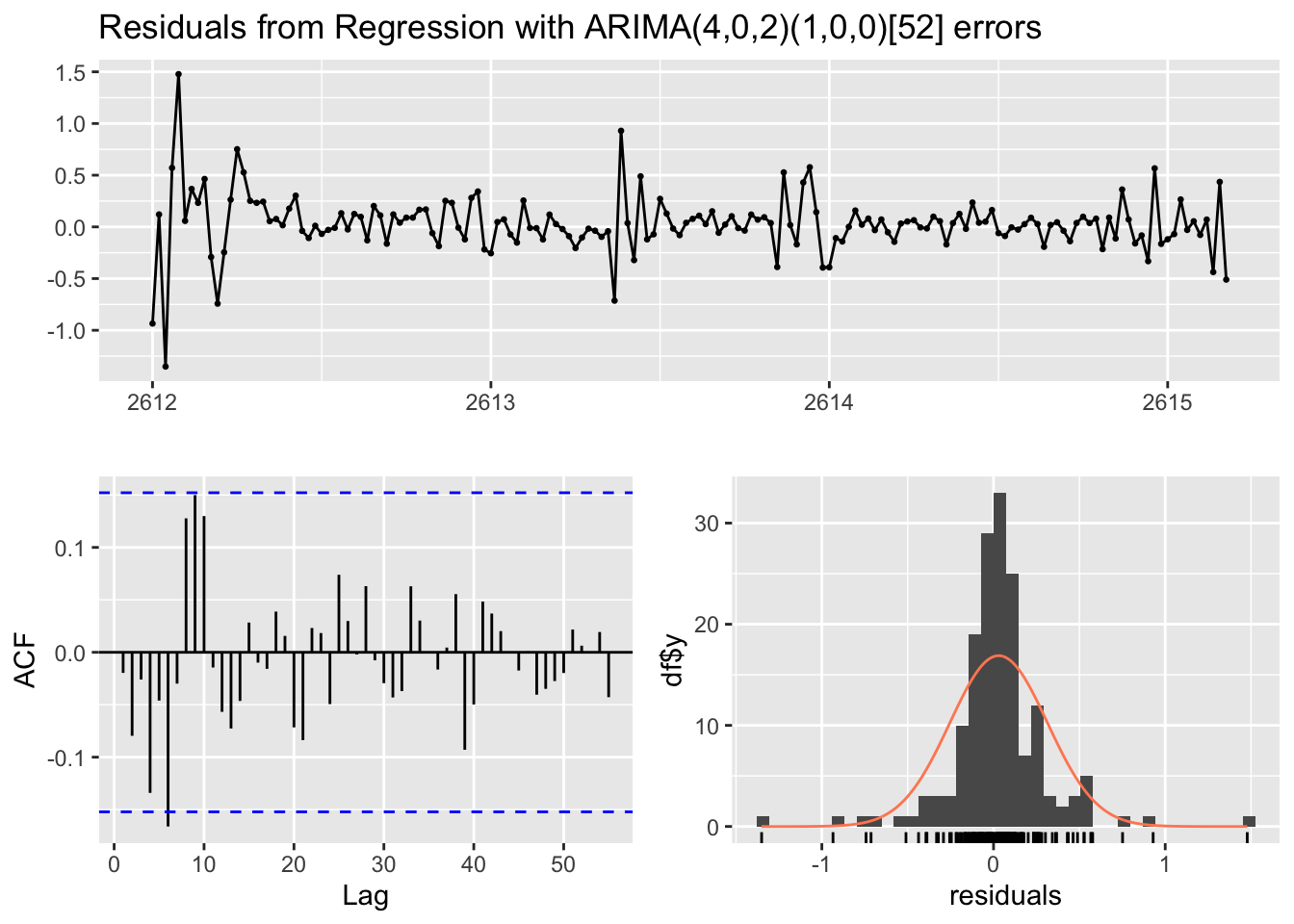
Ljung-Box test
data: Residuals from Regression with ARIMA(4,0,2)(1,0,0)[52] errors
Q* = 28.818, df = 26, p-value = 0.3194
Model df: 7. Total lags used: 33arimax_forecast %>%
residuals() %>%
ljung_box(lag = 10) %>%
knitr::kable()| x | |
|---|---|
| lb_stat | 19.5685060 |
| lb_pvalue | 0.0336084 |
Questions
Q1: Why did we use both visual inspection and formal Chow tests to identify structural breaks?
Q2: Why does weekly aggregation help with structural break detection despite losing temporal granularity?
(Hint: Consider surveillance reporting cycles and weekend effects)
Q3: The original analysis uses vaccine administration as a break source. Propose 3 alternative candidate breakpoints from pandemic history and operationalize their measurement.
Q4: How would you interpret a positive coefficient (0.0182) on the post-vaccine dummy variable?
Q5 : Modify the breakpoint code to test for two or more structural breaks (pre/during/post vaccine eras).
# Solution:
bp_multi <- breakpoints(cases ~ 1 + vaccines, data = covid_weekly, h = 0.02, breaks = 5)
regimes <- covid_weekly$week[bp_multi$breakpoints]
regimes<yearweek[5]>
[1] "2020 W08" "2020 W11" "2020 W43" "2021 W06" "2021 W29"
# Week starts on: Monday\[ \begin{aligned} \log(\text{Cases}_t) &= \phi_0 + \sum_{k=1}^k \gamma_k D_t^{(k)} + \beta_t \log(\text{Vaccines}_t) \end{aligned} \]